from Siebe (@PatientPersists) on Twitter…
Striking claim by @johanvawe at today’s conference:
they found evidence strongly suggesting viral replication (antisense SARS-CoV-2 RNA) via transcriptomics in blood of LC patients!
&
Paxlovid reduced all (viral) biomarkers, except for antisense RNA
This suggests SARS-CoV-2 somehow might evade full shutdown by Paxlovid, and maybe all Mpro inhibitors (like Pax is) need to be coupled with other antiviral therapeutics
The study also found that viral RNA load correlated with patient-reported severity. This adds weight to it playing a causal role
As far as I understand:
Transcriptomics = RNA
Genomics = DNA
Proteomics = proteins
This is the second time strongly suggestive evidence of viral replication has been found.
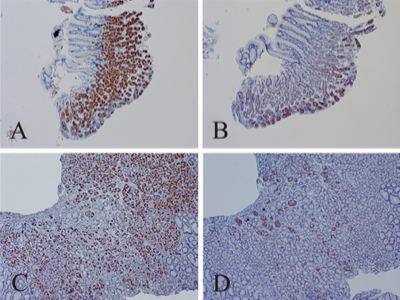
The first time also finding antisense RNA, but in tongue tissue of 4/16 LC patients with taste issues, using in-situ hybridization
https://evidence.nejm.org/doi/full/10.1056/EVIDoa2300046
There are some big open questions:
-
How come other studies didn’t find this?
-
How would SARS-CoV-2 maintain antisense RNA while other proteins are effectively suppressed?
-
Which fraction of patients has replicating virus?
The paper isn’t out yet unfortunately, so we can’t dig into the data yet
Some background info from my amateur understanding:
The target of Paxlovid = Main protease (Mpro/3CLpro/nsp5)
Mpro cuts a huge part of a chain of proteins into single proteins.
Most SARS-CoV-2 antivirals inhibit this Mpro, thereby rendering all those proteins useless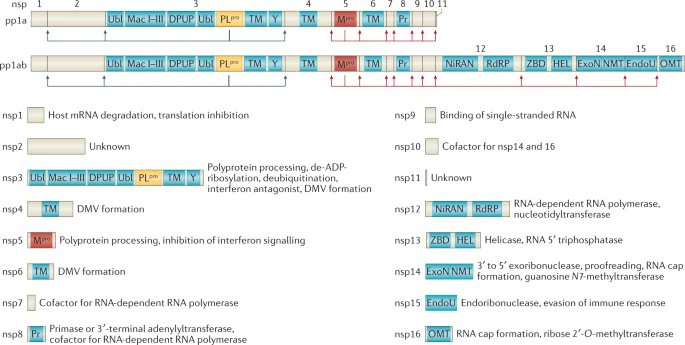
Some of those proteins rendered useless form the RNA-dependent RNA polymerase (RdRp), an enzyme that turns RNA into more RNA
This is absolutely crucial to the viral life cycle.
Sense/positive RNA gets transcribed into antisense/negative RNA, and vice versa
While positive sense RNA has multiple functions, antisense RNA has only 1 function for SARS-CoV-2: serving as a template for positive sense RNA
If you have complete antisense RNA, you have everything needed for viral replication
Antisense RNA is typically much less prevalent than positive sense, for positive single-stranded RNA viruses like SARS-CoV-2, making it harder to detect
It’s commonly said: 100x less prevalent.
Only paper I found on SARS-CoV-2 found a difference 8K-32K!
https://www.sciencedirect.com/science/article/pii/S1201971221000539?pes=vor
I wrote some notes about detecting viral replication with different (highly sensitive methods) here:
I concluded that the most sensitive methods hadn’t been tried yet (branched DNA probes & sequence-specific PCR), but those may now not be necessary
Sensitive detection methods for SARS-CoV-2 in Long Covidhttps://docs.google.com/document/d/1_F5ZhwslH1eWNJ-aS7C030jRPEo74ZP0L5mgEEOLI9o/edit?usp=drivesdk
It is worth noting that in ME/CFS, enteroviruses are suspected to be persisting in a non-cytolytic fashion (i.e. they don’t destroy the host cell), with lower levels of replication, and sense:antisense ratio closer to 1:1
https://me-pedia.org/wiki/Non-cytolytic_enterovirus
Could SARS-CoV-2 become non-cytolytic?
Nobody knows, even though it’s the most studied virus in the world
Coronaviruses are quite different from enteroviruses too
But I did find this paper on a murine CoV that became non-cytolytic (I think?)
Intracellular restriction of a productive noncytopathic coronavirus infection - PubMedVirus infection in vitro can either result in a cytopathic effect (CPE) or proceed without visible changes in infected cells (noncytopathic infection). We are interested in understanding the mechanism…https://pubmed.ncbi.nlm.nih.gov/17959675/
All in all, lots of food for thought!